Fig.7-17 Protocol to analyze DNA strand-break termini using SVPD
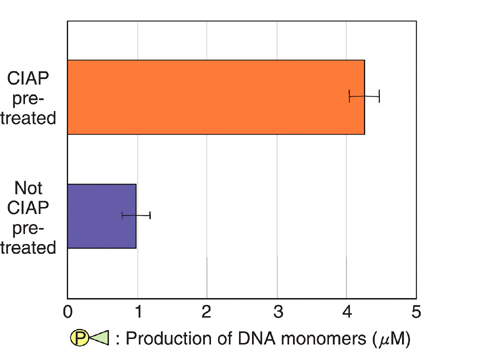
Fig.7-18 Production of DNA monomers from DNA strand-break termini by SVPD
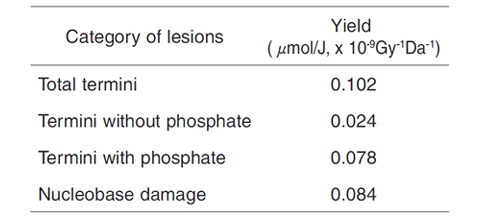
Table 7-1 DNA damage by 60Co γ-rays
Environmental factors stimulating a living body, such as ionizing radiation, can injure DNA which is important for heredity. Ionizing radiation has the ability to cause a variety of types of damage in DNA. This radiation may be classified into γ-rays, X-rays, neutrons, and heavy ion particles. Each type of radiation has an individual DNA damage spectrum. Utilizing this fact, we have developed a new-concept analytical method to estimate the DNA damage spectrum. There are three categories of detection methods so far; one is a method using an antibody recognizing a specific type of damage, the second one uses chemicals reactive to a specific functional group on the damaged site, and the third one uses a supercoiled plasmid DNA. Though these methods are useful to detect damage whose type is known, it is difficult to know the overall spectrum of DNA damage containing undetermined damage. The new method introduced here will be a powerful technique to view overall DNA spectrum.
DNA damage is generally classified into two categories. one is "strand break" , which occurs by cleavage of a molecular chain of DNA. The other is "nucleobase damage" , which occur by destruction of a part of a nucleobase, carrying genetic information of the organism. There are two kinds of termini in the strand break pattern: the termini with or without phosphate (Fig.7-17). We have developed the protocols to quantify (1) termini without phosphate, (2) termini with phosphate, and (3) nucleobase damage. An enzyme, snake venom phosphodiesterase (SVPD), recognizes a terminus without phosphate and cuts DNA monomers sequentially from this terminus. Then, the yield of (1) can be quantified since the amount of the DNA monomers produced during incubation for a given period is proportional to that of damage (1). Furthermore, pre-treatment of irradiated DNA by another enzyme, calf intestine alkaline phosphatase (CIAP), enables damage (2) to be recognized by SVPD since CIAP removes phosphate and so converts damage (2) to damage (1) (Fig.7-17, Fig.7-18). On the other hand, pre-treatment of irradiated DNA by a chemical, piperidine, can covert some of the (3) damage to terminals (2), which can become recognizable after CIAP pre-treatment as mentioned previously (see caption in Fig.7-17). The results obtained from the analysis are shown in Table 7-1. Other methods of DNA damage analysis cannot give us these data in principle. This new analytical protocol is a unique idea, and analyses using a variety of combinations of other enzymes made possible by this methodology will elucidate unknown DNA damage.