Fig.5-11 Protein dynamics during functional expression
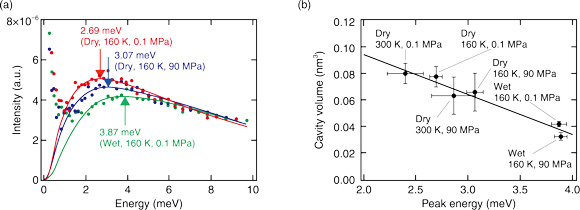
Fig.5-12 (a) Boson peak of a protein (nuclease), and (b) relationship between boson peak position and cavity volume in various environments
Proteins play a central role in all enzymatic reactions from intracellular reactions to chemical reactions catalyzed by enzymes in detergents. Researchers have considered that knowledge of the three-dimensional structure and specific complementing counterpart (substrate), like a lock and its key, is necessary for understanding the function of a protein. However, proteins have a soft structure that fluctuates in response to the surrounding environment; such structural fluctuations are necessary for binding to the substrate molecule (Fig.5-11).
Inelastic neutron scattering enables the quantitative and accurate measurement of structural fluctuations in the THz region (1 THz = 4.1 meV), which is important for the expression of protein functions. The boson peak, which is observed in this energy region, has been demonstrated to be directly related to the structural fluctuation of the protein molecule. Elucidating the relationship between the boson peak of proteins and their three-dimensional structure is vital from the viewpoint of structural science to derive a universal principle for predicting biological reactions from the structural fluctuations of proteins.
In this study, the effect of hydration, temperature, and pressure on a boson peak of the protein (nuclease) were studied by inelastic neutron scattering. The results, as shown in Fig.5-12(a), indicated that hydration, cooling, and pressing each caused the peak position to shift to a higher energy. To investigate the relationship between the peak shift and the protein structure, a molecular dynamics simulation was used to calculate the cavity volume inside the protein. A correlation was found between the peak position and cavity volume (Fig.5-12(b)). Therefore, the fluctuations of the protein structure are related to the atomic packing of the protein structure. Using this correlation between the volumetric properties and structural fluctuations of a protein, measuring the dynamics will help clarify the three-dimensional structure of the protein, even for proteins with unknown structures.
In this study, characteristics of protein dynamics, which are associated with the structure, were elucidated using inelastic neutron scattering and molecular dynamics simulations. This achievement confirms the importance of studying protein dynamics by neutron scattering. Inelastic neutron scattering is expected to become an increasingly effective tool for such studies.
This study was partly supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Challenging Research (Exploratory) (No.18K18828), “Molecular selective phonon spectroscopy at amorphous interface by inelastic neutron scattering”.
(Hiroshi Nakagawa)