When a DNA strand is damaged by radiation, the four bases (T, A, G and C) change chemically and the strand is broken. Almost all damage, however, is restored to the former state due to intracellular repair enzymes. How do the enzymes detect the damage sites in these long DNA molecules? From biological experiments, the following is known: there is a signal on DNA for an enzyme to recognize. The selective binding between DNA and an enzyme takes place due to hydrogen bonds and van der Waals forces between the amino acids of enzymes and bases of the DNA. However, the principle mechanism of selective recognition by enzymes has remained unknown.
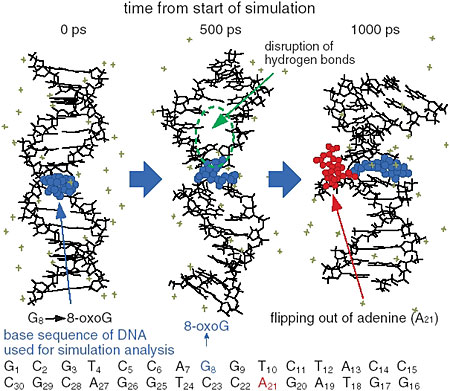
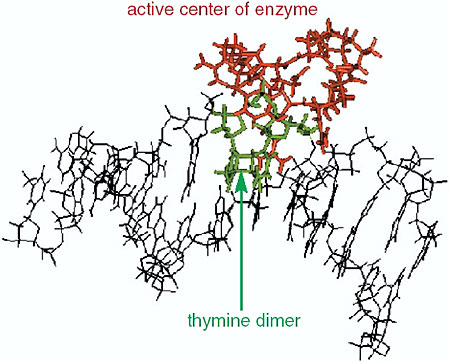
The interaction between DNA base damage and the repair enzyme, which occurs within several thousands of a picosecond, was investigated using molecular dynamics simulation. The thymine dimer (which causes skin cancer) and 8-oxo-guanine (which induces mutation as a carcinogen) were selected as representatives to induce base damage. It was found that 8-oxo-guranine induced a disruption of the hydrogen bond network between complement bases, the double helix opened, and a base in the strand opposite to the damaged strand flipped out, which altered the helical structure (Fig. 3-11). On the other hand, it was found that production of thymine dimer gave rise to a sharp winding in the helical structure of DNA at the center of the damage site (Fig. 3-12), whereas in undamaged DNA, the enzymes did not bound to DNA molecules, as significant structural changes of DNA had not taken place. Therefore, it seems that repair enzymes are able to recognize the helical structural changes of DNA induced around damaged base sites and to form stable complexes with DNA. From the calculation of electrostatic energy changes with the structural change of DNA, it was demonstrated that changes occurred in damage site specific energy. That is, the changes in electrostatic energy at the damage site become a crucial marker for an enzyme to distinguish the damaged site from an undamaged site by scanning DNA molecule surfaces.
|