Fig.10-13 Database for RNA-protein complex structures
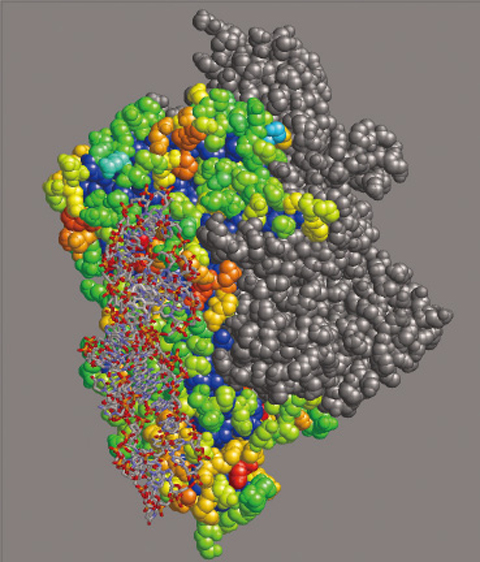
Fig.10-14 A result of RNA interface prediction
Organisms are under constant exposure to radiation including ultraviolet light. Irradiation of organisms is known to damage their DNA. High-energy light as well as oxygen is a known cause of damage to DNA and RNA. The damage is considered to be a cause many biological phenomena including aging. Organisms have evolved mechanisms to repair this damage and maintained the species. Molecules and mechanisms for DNA/RNA repair are the interest of a number of researchers, and extensive studies have been carried out. The studies have shown that protein molecules are the basic molecules for DNA/RNA repair.
DNA sequences of the human genome were all determined at the beginning of the 21st century, and we now have a blueprint for a human. However, the question we face now is how to read the blueprint. DNA sequences of human genome are equivalent to a string of three billion letters written with only four types of characters, and we need to ‘decipher’ the string. The process of deciphering is a collaboration of biological and computational science, and this new field is called bioinformatics. The bioinformatics research explained here is specifically a study to discover a DNA/RNA repair related protein by deciphering DNA sequences.
DNA/RNA repair proteins first need to bind to DNA/RNA. Finding out locations on the proteins for DNA/RNA interactions is one of the first steps for studying DNA/RNA repair. Many researchers have elucidated a lot of structures of protein that bind to a target RNA. We have gathered structural data of proteins and built a database (Fig.10-13). Using this database, we can carry out statistical analysis to find characteristics of RNA interfaces on proteins, such as size of RNA interfaces and characteristic atoms that appear in RNA interfaces. With these statistically derived characteristics, one can perform prediction of RNA interfaces on a protein not yet observed to bind RNA. We are developing new statistical methods and are trying to predict the interfaces with high quality (Fig.10-14).
By predicting RNA interfaces on proteins with high quality, we can design biochemical experiments to determine RNA interfaces. A collaboration of biological and computational sciences is now getting to elucidate mechanisms of molecules that sustain life.