
Fig.10-10 Functions of copper ion in living organisms
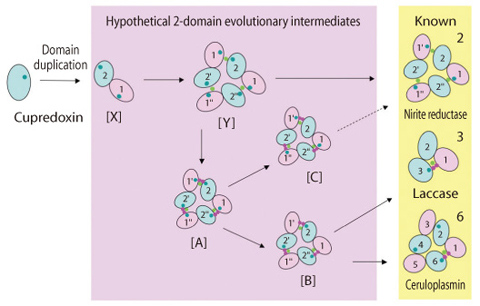
Fig.10-11 Proposed pathway of the evolution of Multi-Copper Blue Proteins
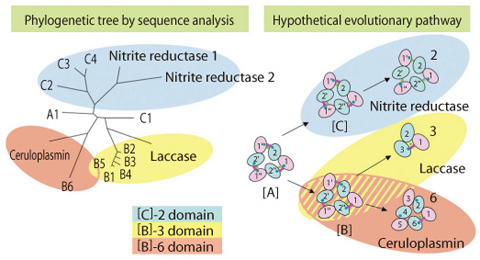
Fig.10-12 Phylogenetic tree and the inferred evolutionary pathway
It has been known that putting 10-yen coin made of copper in shoes prevents foot odor by deterring the growth of bacteria. Copper ion is highly reactive and mediates the formation of active oxygen, which destroys important bio-molecules such as DNA, and therefore is very harmful to simple organisms such as bacteria. However, through the long history of evolution, higher organisms have developed resistance against copper ions, developing a mechanism to excrete the heavy metal and to repair the damaged DNA. Furthermore, some organisms have even acquired a mechanism to exploit the high reactivity of copper ions, for transportation of electron or oxygen, and for the catalysis of the synthesis of useful molecules. (Fig.10-10)
Among those proteins, we focused on a group of proteins called multi-copper blue proteins and carried out analysis. Multi-copper blue protein is a group of proteins which exploits the unique redox ability of copper ions, produced by a wide variety of organisms and having important and versatile functions. It consists of three major groups. 1) Nitrite reductase which is responsible for denitrification, an important process in the circulation of nitrogen in nature. 2) Laccase, responsible for formation/degradation of cell walls of plants/fungi, and ascorbate oxidase which prevents oxidative stress that damages DNA. 3) Ceruloplasmin, responsible for the formation of hemoglobin that carries oxygen in blood stream of vertebrates.
As seen with the completion of human genome project, the amount of nucleotide sequence information is explosively increasing. We used this information efficiently to clarify the evolutionary history of multi-copper blue proteins, (Fig.10-11, Fig.10-12) and discovered new types of multi-copper blue proteins ([A], [B], [C]) from genome sequences.
Furthermore, by carefully comparing those newly identified sequences, we could gain insight into the "molecular evolution" process, through which the proteins have developed new functions.
With more careful analysis of metal-binding proteins and genome sequences, we are aiming to discover new proteins that are related to radiation resistance, such as DNA-repairing enzymes.