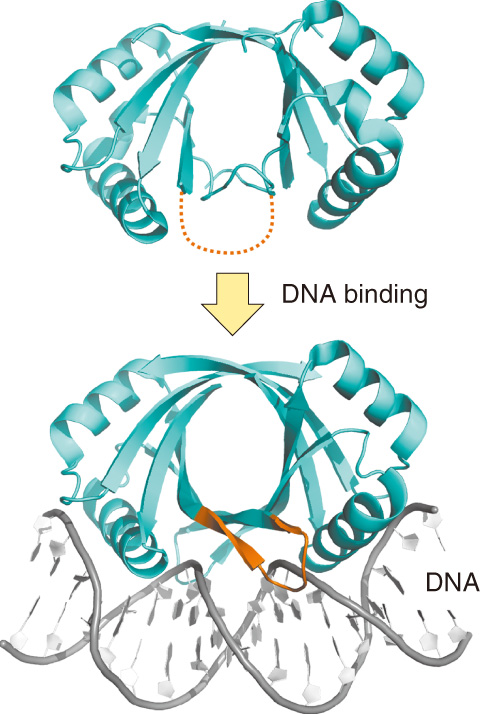
Fig.5-5 Conformational changes in bovine papillomavirus E2 protein upon DNA binding
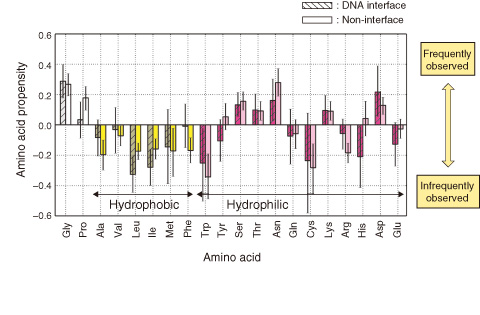
Fig.5-6 Propensities of amino acid residues in conformationally changed fragments
Protein synthesis is controlled by DNA-binding proteins that bind to specific sites in genomic DNA. Therefore, to understand the fundamentals of gene regulation, it is important to elucidate the molecular mechanisms by which DNA-binding proteins recognize specific DNA targets.
Structural studies of DNA-bound as well as DNA-free forms have been conducted to clarify the molecular mechanisms of individual DNA-binding proteins. However, the generic mechanisms of protein-DNA recognition are not yet fully understood.
It is often reported that the DNA interfaces of proteins undergo conformational changes in order to recognize specific DNA targets (e.g., Fig.5-5), but the changes are likely to depend on individual proteins. To assess the structural rearrangement quantitatively, we developed a new method. In this method, protein structures are described as a one-dimensional string of letters. Each letter expresses the conformation of a contiguous four-residue fragment. This enables us to rapidly compare structures and detect the conformational changes in proteins. Using this method, we analyzed a set of proteins whose structures were solved in both DNA-free and DNA-bound forms.
We found that DNA interfaces have higher conformational flexibility than non-interfaces, and that conformationally changed fragments in DNA interfaces have a large amount of glycine, proline, and hydrophilic residues (Fig.5-6). Those residues have previously been observed in intrinsically disordered regions. These characteristics of the amino acids indicate that DNA interfaces evolve to have amino acids that can exhibit high flexibility for accommodating DNA conformations.
DNA recognition by proteins is one of the fundamental processes in gene regulation. Our findings may contribute to the development of a method that accurately predicts DNA binding sites in proteins. Such information can be used to develop drugs that prevent proteins from binding to their target DNA.