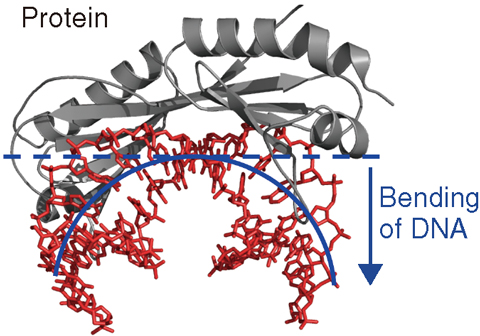
Fig.5-22 Bending of DNA by protein binding
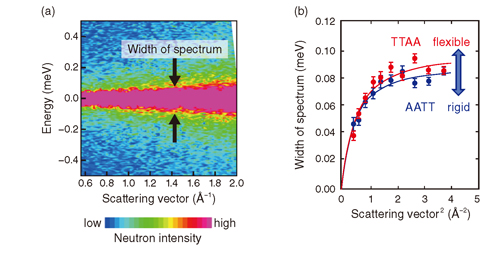
Fig.5-23 (a) Quasi-elastic neutron spectrum of DNA and (b) width of spectrum
The genomes (DNA sequences) of many organisms have been obtained, and a research base for analyzing and predicting biological function from a genome has been established. However, during the process of gene expression, amino acids and bases interact with each other to form a complex tertiary structure of protein and DNA, and a clear correspondence between gene sequence and biological function cannot necessarily be seen. Recent studies have shown that not only is direct interaction between protein and DNA (known as “direct recognition”) important for the information contained with a genome but also is “indirect recognition”, which is an easy transformation of the sequence-dependent DNA structure (Fig.5-22). Because analysis of fluctuations in the sequence-dependent DNA structure under indirect recognition is important, it is necessary to experimentally demonstrate that the dynamics of DNA can be observed directly by experiment.
In this study, we performed quasi-elastic neutron scattering experiments using the AMATERAS instrument at J-PARC/MLF to study the dynamics of two kinds of DNA sequences: 5’CGCGAATTCGCG3’ (AATT) and 5’CGCGTTAACGCG3’(TTAA). These sequences were predicted to be rigid and flexible, respectively, by analysis of a systematic molecular dynamics simulation. We successfully acquired the neutron spectrum to the required precision (Fig.5-23). Having analyzed the quasi-elastic neutron scattering data in detail, it was found that the width of the spectrum was wider with TTAA than with AATT. Thus, this width constitutes a dynamic index by which to observe the difference in the softness of the structure. This result demonstrates agreement with the molecular dynamics simulation and supports the importance of the indirect interaction of DNA flexibility in the recognition of DNA, as has been proposed based on simulations and statistical analyses of crystal structures.
This result demonstrates that DNA sequences should include not only the string of bases but also information about DNA flexibility. We expect that the discovered structural property of DNA in this study will contribute to both elucidation of the mechanism controlling the “on-off switch” of gene-expression and basic research on gene therapy and regenerative medicine guided by cytodifferentiation.
This study was partly supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) KAKENHI Grant-in-Aid for Scientific Research (No.18031042).