![Fig.4-4 Deduced structure of the active site of [NiFe]hydrogenase](img/honbun/e2008_4-4.jpg)
Fig.4-4 Deduced structure of the active site of [NiFe]hydrogenase
![Fig.4-5 Reaction of the model compound of [NiFe]hydrogenase](img/honbun/e2008_4-5.jpg)
Fig.4-5 Reaction of the model compound of [NiFe]hydrogenase
Hydrogenases are bacterial enzymes that catalyze the activation of H2 into two protons (H+) and two electrons (e-). Their structure when they are active in this enzymatic reaction has been attracting much attention since this would elucidate the reaction mechanism. Though a Ni-H-Fe structure, a nickel atom (Ni) and an iron atom (Fe) bridged by a hydrogen atom as shown in Fig.4-4, has been proposed as a possible structure, such a structure has not been observed.
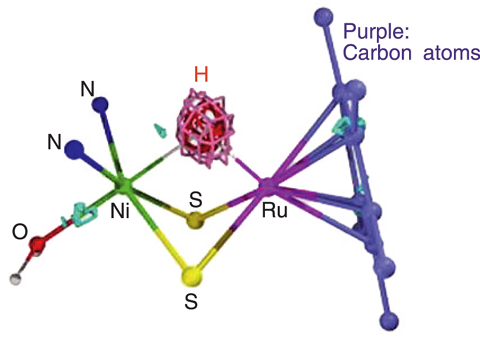
Recently, Prof. Ogo's group at Kyushu University successfully synthesized a model compound with reactivity similar to the reaction center of a [NiFe]hydrogenase, [NiFe]Hase, by using ruthenium (Ru) instead of Fe as shown in Fig.4-5. Whether this compound has a Ni-H-Ru structure or not is important for determining the activation structure of the [NiFe]Hase and to elucidate the mechanism of the enzymatic reaction. However, observation of the hydrogen atom is difficult by the usual structure analysis method, X-ray diffraction, and consequently the Ni-H-Ru structure had not been observed.
Thus, JAEA carried out single crystal neutron structure analysis of this model compound using the "BIX-3" diffractometer at "JRR-3" reactor. A neutron is well scattered by a hydrogen atom and thus a neutron beam is suited for observation of hydrogen atoms. We measured the strength of 10161 diffraction spots in 9 days of measurement and successfully observed the hydrogen atom between the Ni and the Ru atoms as shown in Fig.4-6. Consequently, this model compound was confirmed to be the first compound which has a Ni-H-Ru structure, which indicates that the activation site of the natural [NiFe]Hase has a similar Ni-H-Fe structure.
This result may lead to development of new hydrogen activation catalysts to produce hydrogen energy resources. This research was carried out by Kyushu Univ., JST, Osaka Univ., Univ. of Hyogo and JAEA and published in Science in 2007. JAEA carried out the structure analysis by neutron diffraction.