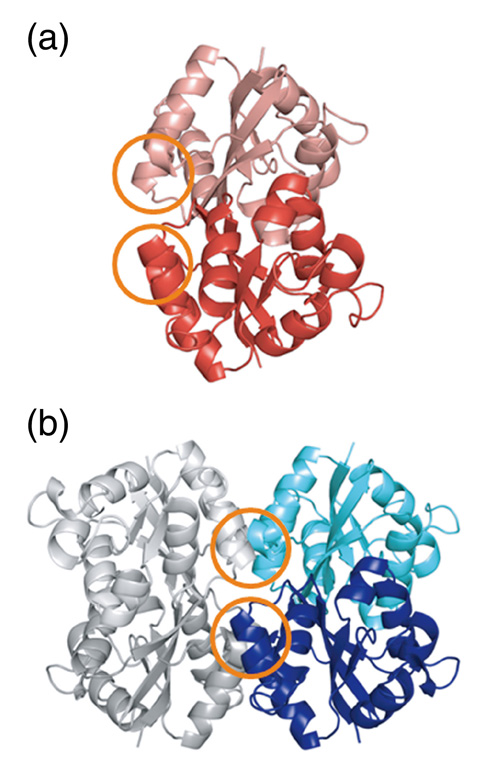
Fig.5-19 Three-dimensional structures HaNDKs
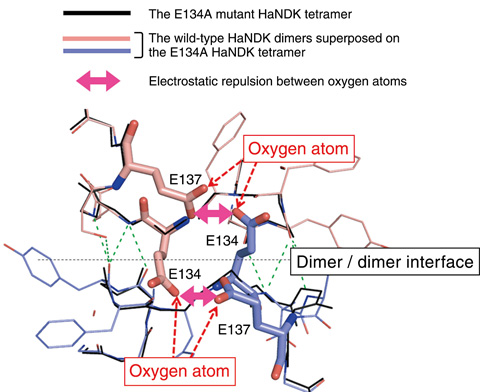
Fig.5-20 The tetramer is the interface newly created by two dimer units of the E134A mutation
In some proteins, the formation of a higher-ordered assembly is essential for enabling their biological function. Such an assembly formation is known to be effective for improving the stability and function of proteins. To develop a technique for operating protein assembly, we attempted to change the assembly state of nucleoside diphosphate kinase (NDK).
NDK exists in the cells of various organisms, and while NDKs from most organisms assemble into a tetramer or a hexamer, NDK from the halophilic bacteria Halomonas sp.593 (HaNDK) exceptionally forms a dimer. To identify the reason for the lack of dimer-dimer association of wild-type HaNDK, its three-dimensional structure was determined via X-ray crystallography (Fig.5-19(a)). It was found that, when the structure of the wild-type HaNDK was superposed on the structure of a known tetrameric NDK, the 134th and 137th glutamic acids (E134, E137) directly contacted the E134 and E137 positions in the neighboring HaNDK molecule (Fig.5-20). The fact that the overlay provides at least four negative charges at the interface of the two dimer units suggests that the electrostatic repulsion prevents the dimer-dimer association.
Given these results, we expect that the HaNDK dimer will be associated into a higher-order assembly form if the E134 glutamic acid is replaced with alanine (E134A), which has an uncharged short side chain. In fact, the mutation of E134 to alanine (E134A mutant HaNDK) resulted in conversion of the assembly state of HaNDK from a dimer to a tetramer, as shown in (Fig.5-19(b)). Moreover, the specific activity of the HaNDK tetramer was 19% higher than that of the HaNDK dimer.
This study demonstrated that the assembly state of a protein can be controlled with a minimum mutation if detailed structural information is available. The investigation of the structural differences in the dimeric and tetrameric assemblies will provide useful information for creating higher-ordered assemblies, which will contribute to the creation of proteins with increased enzymatic activity and stability.
Our research was accomplished with the support of Kagoshima University.