Fig.4-18 Positions of all atoms in HIV-1 protease-inhibitor KNI-272 complex
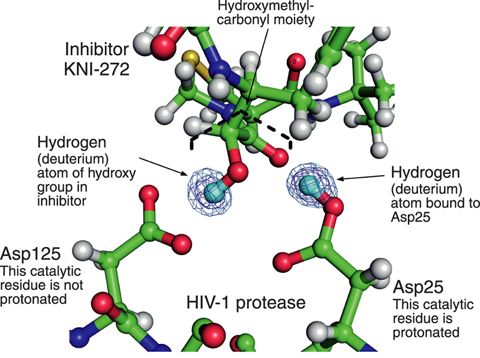
Fig.4-19 Structure in active site of HIV-1 protease
We have succeeded in determining the positions of all the atoms of an HIV-1 protease (protein)-KNI-272 (drug candidate) complex, including hydrogen atoms, by neutron crystallography.
HIV-1 protease is a dimeric aspartic protease which contains two catalytic aspartic residues (Asp25 and Asp125, defined by residue positions), and which plays an essential role in viral replication. To develop structure-based drug designs for HIV-1 protease inhibitors, it is necessary to understand the catalytic and inhibitor recognition mechanisms of HIV-1 protease. Many structural analyses of HIV-1 protease using X-ray diffraction have been reported. However, hydrogen atoms which are important for various functions have not been observed because the scattering factor of hydrogen atoms is much lower than that of other atoms. Thus, we detected those functional hydrogen (deuterium) atoms using neutron quantum beams which are effective for this purpose.
Neutron diffraction data was collected on the BIX-4 diffractometer installed at the 1G-A port of the research reactor JRR-3 in JAEA Fig.4-18 shows the positions of all the atoms, including hydrogen atoms, of the HIV-1 protease - KNI-272 complex.
Inhibitor KNI-272 used in this study contains a hydroxymethyl-carbonyl moiety that interacts with catalytic residues of HIV-1 protease. The neutron diffraction analysis directly showed that Asp25 is protonated and that Asp125 is non-protonated (Fig.4-19). Although the catalytic mechanism of HIV-1 protease has been a matter of some debate (due primarily to lack of direct visualization of hydrogen atoms in the catalytic mechanism), our results demonstrate that Asp25 provides a proton to the carbonyl group of the substrate and Asp125 contributes by activating the hydrolyzing water molecule for nucleophilic attack.
The structural information obtained in this study, including the protonation state of the catalytic residues, will provide important information for designing more effective inhibitors.
This study was performed in collaboration with Kyoto Pharmaceutical University, Osaka University and SOSHO, Inc. and published in Proceedings of the National Academy of Sciences of the United States of America.