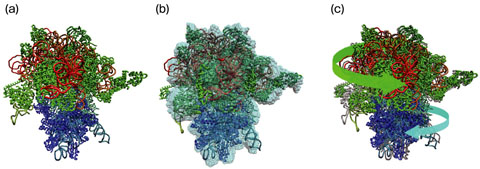
Fig.4-24 Three-dimensional structures of a ribosome determined by different methods
Due to the emergence of more and more antibiotic-resistant bacteria, the development of new antibiotics is an urgent necessity. Different antibiotics attack bacteria in different ways. Some kinds of antibiotics inhibit the growth of bacteria by preventing their protein synthesis. In a cell, protein synthesis is performed by huge macromolecular complexes called ribosomes. For effective development of antibiotics, it is important to reveal the three-dimensional (3D) structure of ribosomes in functional states at the atomic level.
The atomic structure of a biological molecule such as protein and DNA is usually determined by a method called X-ray crystallography. This method requires the crystallization of the molecule, and for the crystallization, many conditions must be satisfied. Thus, it is not easy to determine the structure of the molecule in a specific functional state, and it is especially difficult for a huge molecular complex like a ribosome. The atomic structures of ribosomes in a few functional states have hitherto been determined (Fig.4-12(a), (c)), but this is not enough for understanding the whole molecular mechanism.
In recent years, the use of an electron microscope (EM) has been a popular method for determination of the 3D structure of biological molecules. In this method, the 3D structure is reconstructed from many EM images of the molecule taken from different angles. The resolution of the reconstructed structure is too low to identify the constituent atoms. However, because crystallization is unnecessary, determination of the 3D structure is relatively easy, and the 3D structures of ribosomes in many functional states have already been determined (Fig.4-12(b)).
We have developed a computational technique for building a molecular model from a 3D-EM structure (Fig.4-12(b), (c)). In this technique, the atomic structure determined by X-ray crystallography is deformed so that it fits into the 3D-EM structure. Thus, using this technique, we can determine the atomic structure of a molecule in a specific state, where the structure is known from EM, but not from X-ray crystallography. We have applied this technique to many 3D-EM structures of ribosomes and built molecular models. By analyzing these models statistically, we were successful in clarifying conformational change in ribosomes during protein synthesis.
The structural information that we obtained in this study will contribute to the development of new antibiotics.